Biomedicines, Free Full-Text
Por um escritor misterioso
Descrição
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) caused a global pandemic of coronavirus disease in 2019 (COVID-19). Genome surveillance is a key method to track the spread of SARS-CoV-2 variants. Genetic diversity and evolution of SARS-CoV-2 were analyzed based on 260,673 whole-genome sequences, which were sampled from 62 countries between 24 December 2019 and 12 January 2021. We found that amino acid (AA) substitutions were observed in all SARS-CoV-2 proteins, and the top six proteins with the highest substitution rates were ORF10, nucleocapsid, ORF3a, spike glycoprotein, RNA-dependent RNA polymerase, and ORF8. Among 25,629 amino acid substitutions at 8484 polymorphic sites across the coding region of the SARS-CoV-2 genome, the D614G (93.88%) variant in spike and the P323L (93.74%) variant in RNA-dependent RNA polymerase were the dominant variants on six continents. As of January 2021, the genomic sequences of SARS-CoV-2 could be divided into at least 12 different clades. Distributions of SARS-CoV-2 clades were featured with temporal and geographical dynamics on six continents. Overall, this large-scale analysis provides a detailed mapping of SARS-CoV-2 variants in different geographic areas at different time points, highlighting the importance of evaluating highly prevalent variants in the development of SARS-CoV-2 antiviral drugs and vaccines.

Biomedicines, Free Full-Text

Catalytically Active Snake Venom PLA2 Enzymes: An Overview of Its Elusive Mechanisms of Reaction
Biomedicines, Free Full-Text
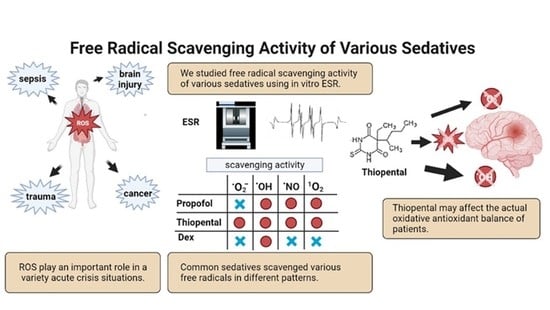
Free Radicals Brain Injury - Colaboratory

Archives Biomedicine
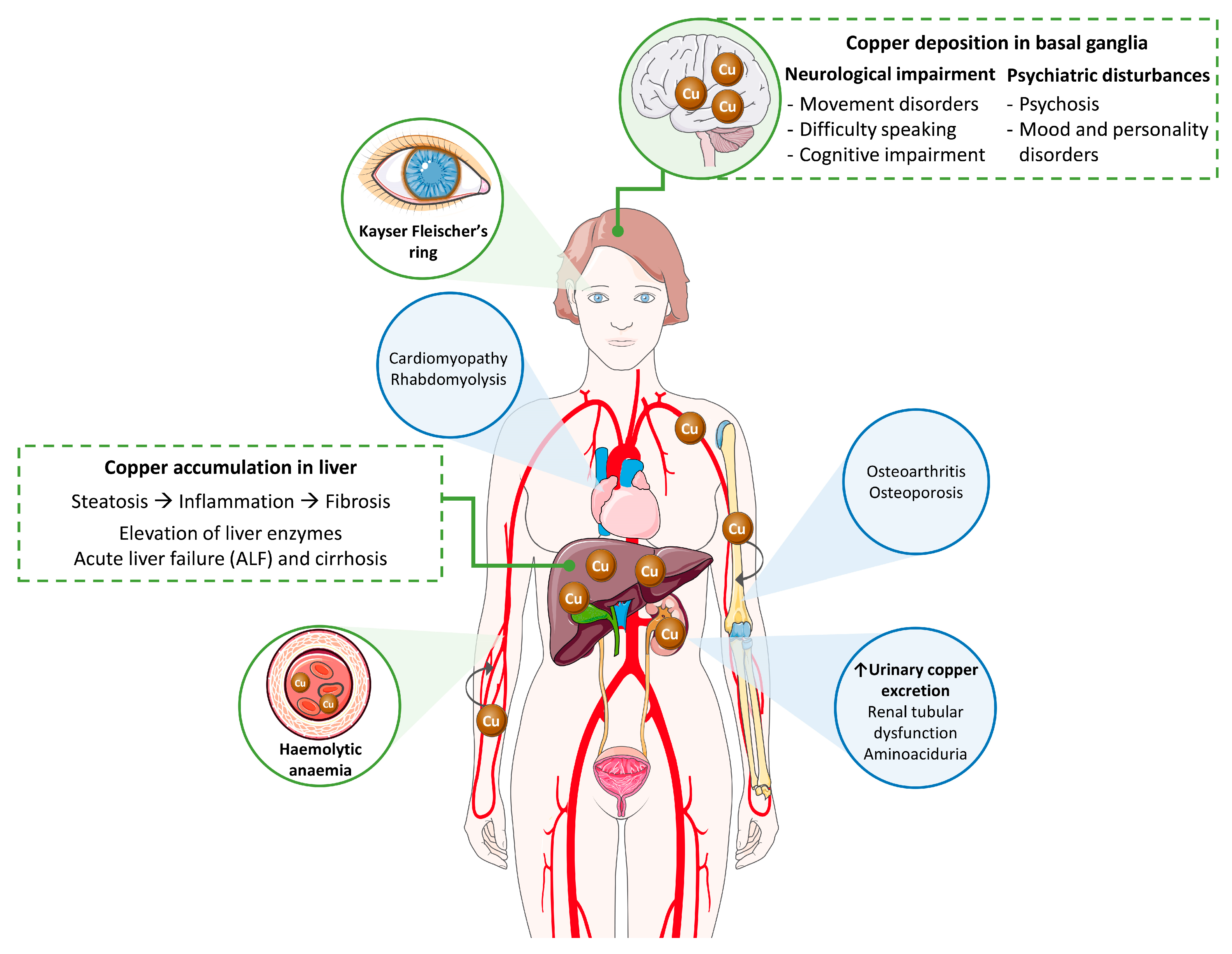
Biomedicines, Free Full-Text
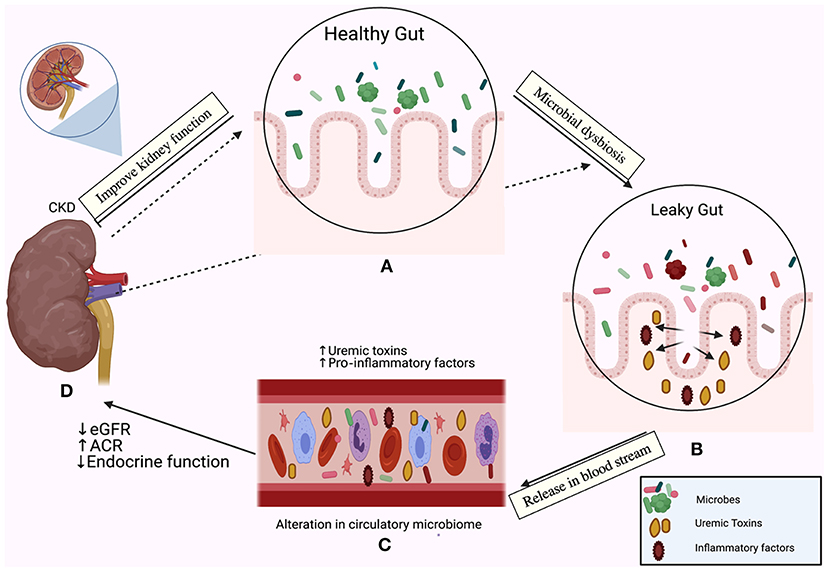
Frontiers The Human Microbiome in Chronic Kidney Disease: A Double-Edged Sword

Biomedicine & Pharmacotherapy, Journal

The Ageless Generation: How Advances in Biomedicine Will Transform the Global Economy: Zhavoronkov, Alex: 9780230342200: : Books

Generate Biomedicines Company Profile: Valuation, Funding & Investors
de
por adulto (o preço varia de acordo com o tamanho do grupo)