Identification of levoglucosan degradation pathways in bacteria and sequence similarity network analysis
Por um escritor misterioso
Descrição
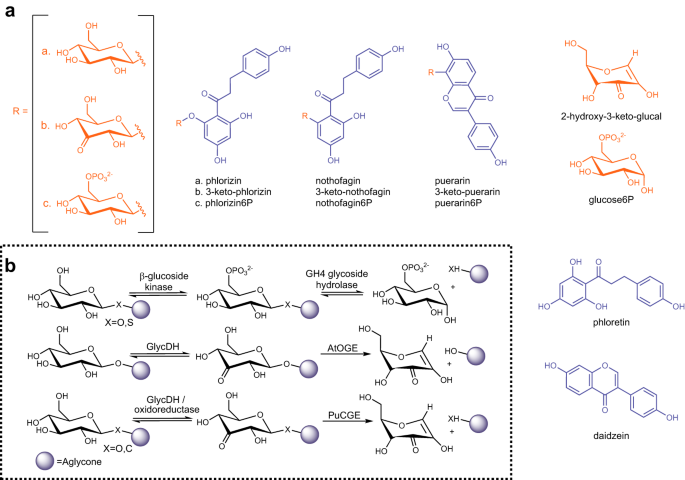
Enzymatic β-elimination in natural product O- and C-glycoside deglycosylation

FAD-dependent C-glycoside–metabolizing enzymes in microorganisms: Screening, characterization, and crystal structure analysis

Isolation and Characterization of Levoglucosan-Metabolizing Bacteria
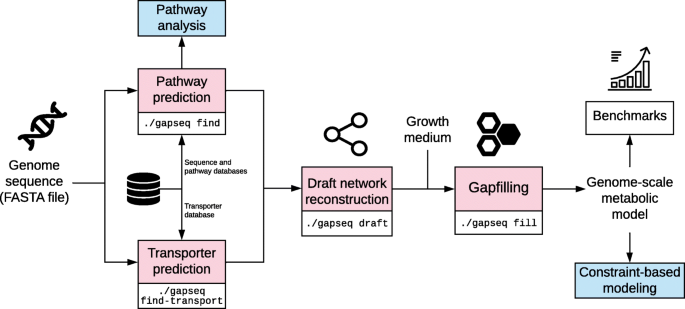
gapseq: informed prediction of bacterial metabolic pathways and reconstruction of accurate metabolic models, Genome Biology
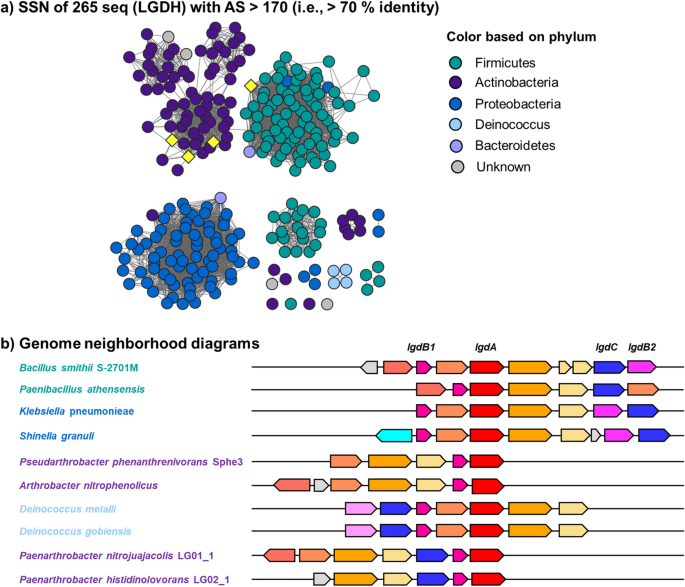
Identification of levoglucosan degradation pathways in bacteria and sequence similarity network analysis
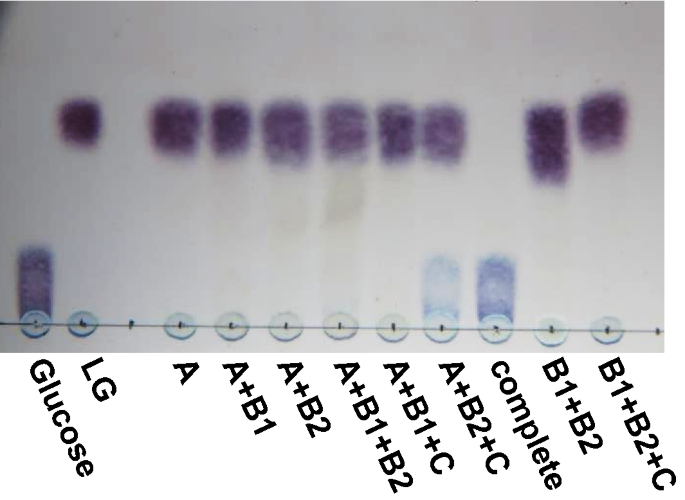
Conversion of levoglucosan into glucose by the coordination of four enzymes through oxidation, elimination, hydration, and reduction

Inhibitor tolerance and bioethanol fermentability of levoglucosan-utilizing Escherichia coli were enhanced by overexpression of stress-responsive gene ycfR: The proteomics-guided metabolic engineering - ScienceDirect

Inhibitor tolerance and bioethanol fermentability of levoglucosan-utilizing Escherichia coli were enhanced by overexpression of stress-responsive gene ycfR: The proteomics-guided metabolic engineering - ScienceDirect

Production of levoglucosan (LG) by pyrolysis of cellulose, and
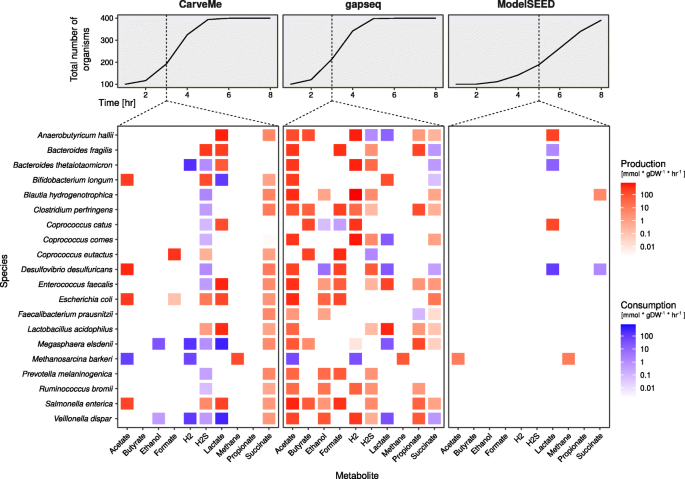
gapseq: informed prediction of bacterial metabolic pathways and reconstruction of accurate metabolic models, Genome Biology

Strain Identification and Quantitative Analysis in Microbial Communities - ScienceDirect

Isolation and Characterization of Levoglucosan-Metabolizing Bacteria

FAD-dependent C-glycoside–metabolizing enzymes in microorganisms: Screening, characterization, and crystal structure analysis

Impacts of Chemical Degradation on the Global Budget of Atmospheric Levoglucosan and Its Use As a Biomass Burning Tracer
Publications - Scott lab
de
por adulto (o preço varia de acordo com o tamanho do grupo)